Axe Architecture du noyau et oncogènes – Equipe B-NATION
Présentation
Team B-NATION is composed of 4 permanent researchers and benefits from technical support and expertise in microscopy, flow cytometry and transgenic mouse work. The team is training post-docs and PhD students, and regularly welcomes Master trainees, Licence and BTS students.
Our team has been focused on cutting-edge basic research on B cell genetics “how does it work?”. B-NATION studied regulation and dysregulation of immunoglobulin gene expression and DNA remodelling with traditional but unique mouse models (developed in the past two decades) and innovative molecular biology tools. More recently, the team has become interested in genome organization and nuclear architecture and is revisiting, at this scale, immunoglobulin gene and oncogenes loci regulation in B-lineage cells.
Development of highly precise and up to date tools (high throughput DNA and RNA sequencing, even now at the single cell level) encourages us to revisit our models in order to refine previous hypotheses. Emerging strategies have developed to study chromosome and chromatin conformation to study locus-wide (and possibly inter-loci) cross-talk between distant cis-regulatory elements. Beyond the enormous interest in physiology, such approaches might provide useful insights into the mechanisms that mediate oncogene translocations and deregulations.
Team members are part of the Cancéropôle GSO network (involved in the steering committee of Genome Dynamics and Cencer axis), the Oncosphere Nouvelle Aquitaine network (EP in the steering committee), members of the Société Française d’Immunologie and part of its Germinal Center Club. EP is a member of the American Association of Immunologists. SLN is a member of CNRS National Committee (section # 27).
Thématiques de recherche
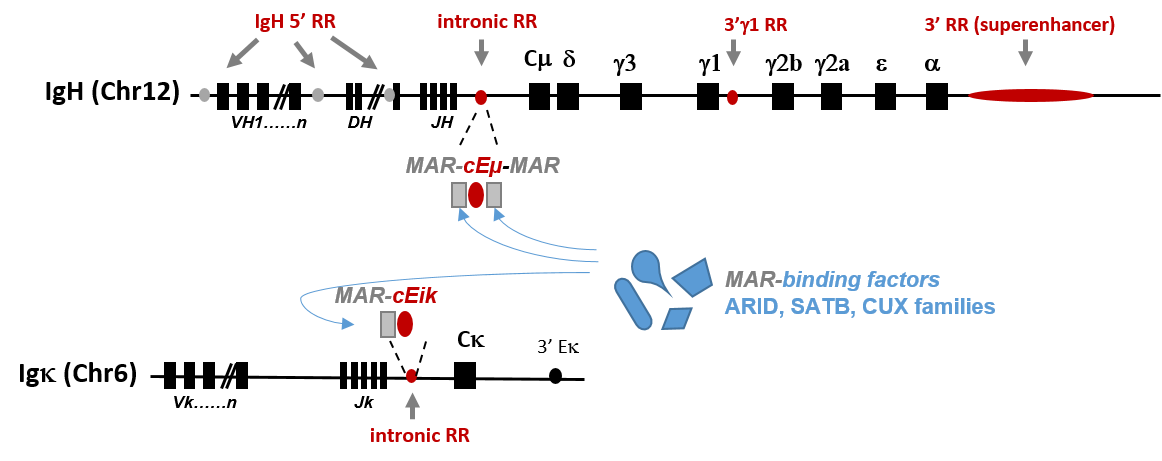
In Axis 1 we continue to study IgH regulatory elements and remodelling events, based on multiple-targeted loci of mouse models. Our projects intend to focus on main and newly described regulatory regions on both Immunoglobulin Heavy and Light chain loci.
Our team also focuses on the function of Nuclear Scaffold/Matrix Attachment Regions (MARs) that either constitutively border chromatin domains (constitutive MARs) or bind tissue-specific nuclear factors that participate in gene regulation expression (facultative MARs). Taking advantage of our mouse models that delete IgH intronic regulatory elements (cEμ or MAREμ) and based on our recent data highlighting a function for MARs in SHM, we intend to study some of the molecular mechanisms that physiologically enhance and limit somatic hypermutation to specific Ig gene regions and loci.
The group also acquire mouse models dedicated to studying proteins interacting with the B cell nuclear matrix (such as the AT-rich binding factors of ARID, SATB and CUX families) expressed in the B-lineage cell and studies the impact of such factors on B cell remodeling events.
In Axis 2 we study the potency of Ig gene regulatory elements to modify nuclear organization of the B cell genome. Our team intends to give high priority to this emerging topic: nuclear architecture. Most Ig gene regulation studies have been so far performed at the nucleosomal scale (genes, loci and epigenetic modifications). Now, increasing interest for the understanding of gene regulation at the whole nuclei scale makes it necessary to revisit the previous models at both the supranucleosomal scale (DNA loops and Topologically Associated Domains: TAD) and the nuclear scale (chromosome territories and nuclear positioning).
Our team is investigating the implication of multiple Ig loci regulatory regions/elements on global genome organization. For this, we setup several tools and methods dedicated to studies at the whole nucleus scale: visual nuclear imaging (3D-DNA FISH method to nuclear distance measurements) and high throughput methods to detect/quantify long range chromosomal interactions (3C-HTGTS), etc…
In Axis 3 we study oncogenic effects of Ig gene regulatory elements and some their associated factors on mutations to oncogenes and illegitimate/atypical rearrangements with the goal to decipher some fundamental pathways linking their implication in CSR/SHM/DNA repair -associated events during translocations and lymphomagenesis. For this, the team takes advantage of several of tools developed in the past decades: animals models for lymphomagenesis, a bioinformatic pipeline dedicated to the detection of somatic hypermutations in Ig (frequent) and non-Ig (rare) genes (Martin et al. J Immunol 2018) , a high-throughput method named LAM-HTGTS that detects genome rearrangement events including translocations.